Genome of golden algae decoded

The Oder disaster in 2022 was man-made, but the direct cause was the toxin produced by a microalgae. Researchers have now been able to identify the genes responsible for the toxin formation.
07/22/2024 · News · Leibniz-Institut für Gewässerökologie und Binnenfischerei · Umweltwissenschaften · Forschungsergebnis
In the summer of 2022, around 1,000 tonnes of fish, mussels and snails died in the River Oder. Although the disaster was man-made, the immediate cause of death was the toxin of a microalgae with the scientific collective name Prymnesium parvum, often referred to as 'golden algae'. Since then, these unicellular organisms have colonised the Oder permanently. A research team led by the Leibniz Institute of Freshwater Ecology and Inland Fisheries (IGB) has now sequenced the complete genome of the microalga in order to identify future risk factors under which the alga multiplies and produces its toxin. They were able to identify the gene sequences that encode the toxins an important step toward an early warning system. The study was published in the journal Current Biology.
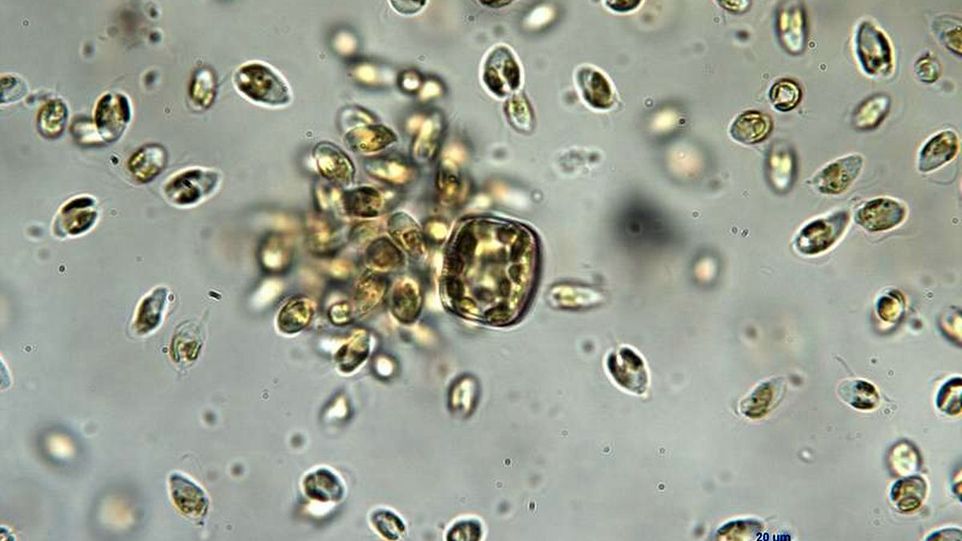
Prymnesium parvum s.l. (sensu lato), commonly known as 'golden algae', stands for a whole group of microalgae which, although tiny at 5 to 10 micrometres in size, can cause devastating damage to ecosystems. This is because these algae can produce cytotoxins, so-called prymnesins. These destroy the gills of fish and filter-feeders such as mussels and snails and also attack other body tissues. The result: death due to lack of oxygen or circulatory failure.
Not all microalgae are the same
Previous studies on morphology and genetics have shown that Prymnesium parvum s.l. exhibits great diversity: it is a complex of at least 40 genetically distinct strains that differ in genome size and produce type-specific prymnesins as well as strain-specific mixtures of different prymnesin variants. Depending on toxin production, three clades are distinguished: A, B or C. To date, there has only been one reference genome, that of type A.
Close relationship between the microalgae ODER1 and brackish water strains from Denmark and Norway
As part of the ODER~SO project, an international team led by IGB researchers Dr Heiner Kuhl, Dr Jürgen Strassert, Prof Dr Michael Monaghan and PD Dr Matthias Stöck has now sequenced the entire genome of the Prymnesium parvum strain from the Oder catastrophe and identified gene sequences that are responsible for the chemical structure of the toxins and thus for their properties. The sequenced strain was named ODER1 and is a member of clade B.
The researchers also created a phylogenetic tree of various Prymnesium parvum strains. This shows that the ODER1 strain is most closely related to another type B strain, K-0081, which was isolated from brackish water in north-west Denmark in 1985, as well as to other type B strains from Norway (RCC3426, KAC-39 and K-0374). This similarity is due to the geographical proximity, but does not provide any direct information on how the alga reached the Oder.
Reference genome for monitoring algal blooms
Following the decoding of a type A reference genome and now the type B reference genome, two very different microalgae of the group have been covered; the decoding of the type C reference genome is still pending. "The decoding of the second reference genome of Prymnesium parvum s.l. provides important insights into the genetic basis and structural variability of the toxins. It has recently been shown that the prymnesin type influences toxicity. This means that we can now estimate the potential toxicity of future algal blooms much better", said IGB researcher Dr Jürgen Strassert, co-author of the study.
Next step: developing molecular methods of toxin analysis and investigating influencing factors
At present, toxin formation cannot be monitored directly. The toxin becomes too diluted to measure in the water and there are currently no standard methods, not even for clade A. "One of the IGB team's next steps will be to analyze toxin formation at the molecular level by determining the expression of specific toxin synthesis genes", added IGB researcher Dr. Heiner Kuhl, lead author of the study.
Environmental conditions play an important role in both the proliferation of the algal bloom and the production of toxins. "Decoding the genes for toxin production is therefore crucial for analyzing the environmental conditions under which the algae form these blooms and possibly produce specific toxins in different amounts", said IGB researcher Matthias Stöck, who led the study.
Further information and contact
Presse release - Leibniz Institute of Freshwater Ecology and Inland Fisheries (IGB)Outlines | IGB Fact Sheet: State of knowledge on the toxic alga Prymnesium parvum in the Oder River